

[Presented by: Andrew Chapman (andrewgchapman@gmail.com), Andrea Deazley (smarty2803@hotmail.com) and Calan Fisher (calan46@yahoo.com). November 6, 2007]
 |
[Presented by: Andrew Chapman (andrewgchapman@gmail.com), Andrea Deazley (smarty2803@hotmail.com) and Calan Fisher (calan46@yahoo.com). November 6, 2007]
|
Introduction
The biological activities of each cell are largely determined by the active proteins expressed in it.
But in cells, some proteins are found in very low quantities, and others are quite abundant, so how is this protein expression controlled? --- Regulation of Gene Transcription
Transcriptional regulation is more accessible than regulation at the other steps along the DNAà RNAà Protein pathway since transcription is the first step of information transfer.
Genome’s contribution to transcription: parts list and instruction manual.
Logic of Prokaryotic Gene Regulation
A cell needs a different enzyme to transport each molecule into the cell, and yet another to breakdown or change each of these molecules into something usable to the cell.
If the cell simultaneously and constantly produced every enzyme it could possibly need, it would take more energy to produce all these enzymes than what would be derived from breaking down the molecules.
For this reason, cells have development mechanisms to repress the genes which encode for enzymes/proteins that are not needed at a given time, but still be able to activate these genes when needed.
The Basics of Prokaryotic Transcriptional Regulation
Prokaryotic transcription regulation requires a series of DNA-protein interaction at sites near the beginning of a gene.
The promoter and RNA polymerase interact to begin transcription a few bases away from the promoter sequence
A gene must have a promoter to be transcribed
There is another DNA-protein interaction which occurs, and this determines if transcription will continue
In this interaction regulatory proteins interact with the operator (a segment of DNA near the promoter) in either positive or negative regulation.
Regulation of the Lactose System
The lac Structural Genes
The lac regulatory components
In addition to RNA polymerase, there are 3 important components in the basic regulation of the lac operon:
1) The Repressor gene – I – which encodes a lactose repressor protein which blocks the expression of the structural genes
2) The lac promoter site – where RNA Polymerase binds to initiate transcription
3) The lac operator site – O – which is located between the promoter and the Z gene; this is where the repressor protein binds.
The lac Operon

Catabolite Repression of the lac operon
The cell can capture more energy from the breakdown of Glucose than Lactose.
If both sugars are present the synthesis of β-galactosidase is not induced until the Glucose is metabolized.
A breakdown product of glucose prevents activation of the lac operon by lactose (catabolite repression)
This catabolite modulates the level of cyclic adenosine monophosphate (cAMP)
When Glucose concentration is high cAMP concentration is low and vice-versa.
cAMP binds allosterically to CAP (catabolite activator protein)
This complex can bind to CAP site on the promoter increasing affinity of RNA polymerase so the lac genes can be transcribed

Results of Genetic Analysis
Constitutive I- mutations- the DNA binding site of the repressor protein has been mutated so that it is unable to bind to the operator
Constitutive OC mutations- The operator has been been mutated so that it can no longer bind the repressor
Genetic Evidence for Allostery
The lac repressor inhibits transcription of the lac operon in the absence of an inducer, and allows transcription in the presence of an inducer. This occurs due to the allosteric site on the repressor. This site binds to the inducer and when bound the structure of the DNA-binding site is changer so that it is no longer able to bind to the operator
Is repressors (super-repressors) cause repression even in the presence of an inducer as seen when comparing strains 1 and 2
Dual Positive and Negative control: The Arabinose Operon
Metabolic Pathways
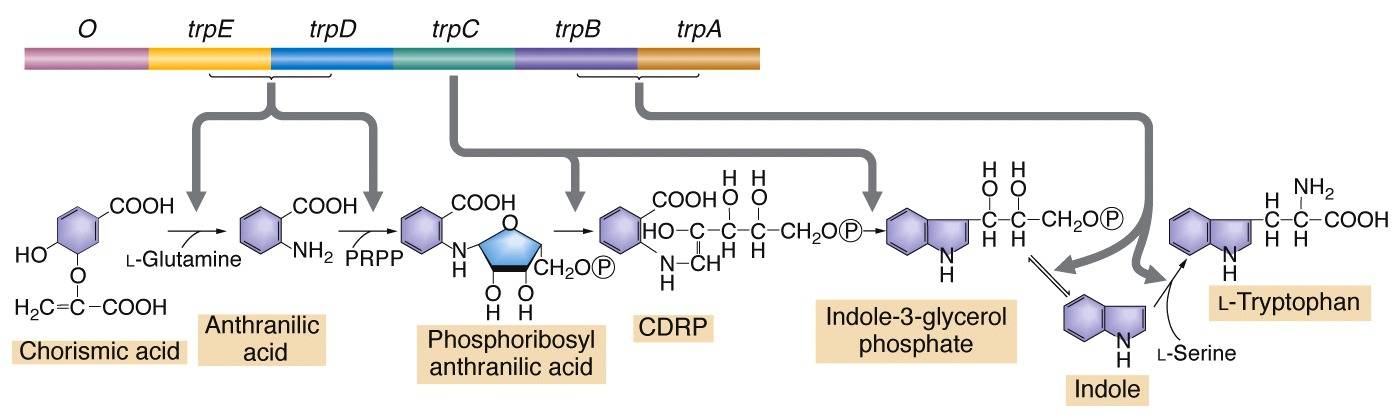
Cis-Acting Sequences in transcriptional regulation
Trans Control of Transcription
There have been a large number of trans-acting regulatory proteins identified in eukaryotes.
These act by binding to specific target DNA sequences-namely the promoters, promoter-proximal elements, and distance-independent elements
Most regulatory proteins bind to core promoter and promoter-proximal elements. These help RNA polymerase II initiate transcription by forming an initiation complex with the polymerase.
The proper configuration of proteins binding to the core promoter, the promoter-proximal elements, and distance-independent elements is required for the effective initiation if transcription.