inverted terminal repeat - dsDNA reads the same in both orientations
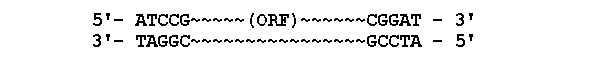
=> elements may move from one position to another in genome
The genome is not
static: genes move
Phenomenon
identified in 1940's by B. McClintock
in maize: Nobel Prize 1983
[Keller: "A Feeling for the Organism"]
Ac
- Ds system in maize (Zea)
Ds
(Dissociator) locus causes chromosome breaks
only when Ac (Activator) locus is present
in trans configuration
Break affects expression
of other genes in cis configuration
Ds
element may "jump" elsewhere in genome:
effect on expression of other genes is unstable,
depending on position of Ds
Ac
& Ds are transposable elements
inverted terminal repeat - dsDNA reads the same in both
orientations
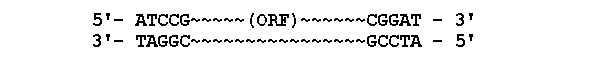
=> elements may move from one position to another in genome
Ac
has an ORF for a presumptive 'Transposase'
gene
Ds is Ac with a defective Transposase
gene
Do transposable elements account for 'classical' mutants?
'wrinkled' (rr)
peas (Pisum)
defect of starch-branching enzyme (SBEI)
'white eyes' in Drosophila
insertion of copia sequences
Alu
sequences in Homo
300 bp element, 9 x 105 copies => 10% of human genome
is mobile