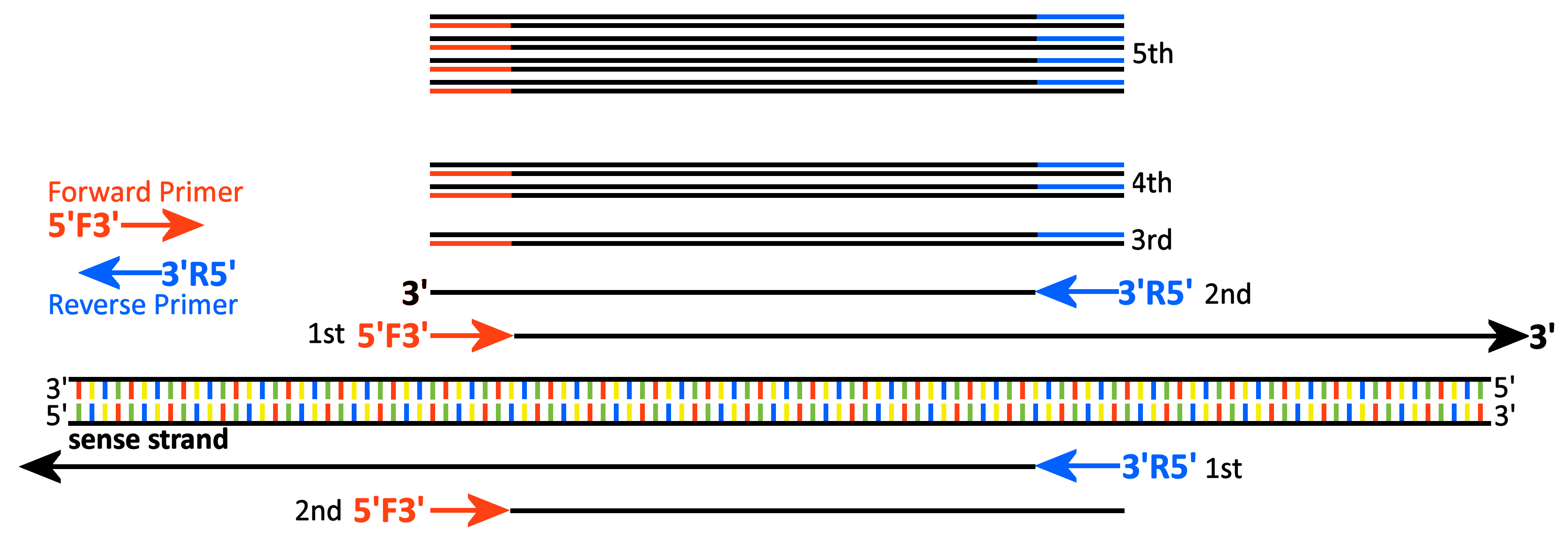
The Polymerase Chain Reaction (PCR) in detail
A piece of dsDNA includes the target region of
interest [middle, base-paired DNA]. We draw it such that the Sense
Strand is on the bottom, with the 5' 3' direction drawn left to right.
The reverse complement Partner Strand is drawn on the
top, with the 3'
3' direction drawn left to right.
The reverse complement Partner Strand is drawn on the
top, with the 3' 5'
direction drawn right to left.
5'
direction drawn right to left.
The first denaturation step separates the two strands. The first Annealing step allows the Forward (5'F3') and and Reverse (3'R5') Primers to anneal to their complementary sequences in the top and bottom strands, respectively. The primers are on opposite strands at either end of the intervening target region. The 5' 3' orientation of the Primers is always anti-parallel
to the target strand 3'
3' orientation of the Primers is always anti-parallel
to the target strand 3' 5' as
drawn. The first Extension step extends both primers in the 5'
5' as
drawn. The first Extension step extends both primers in the 5' 3' direction. Both produce 'long
products' as they extend indefinitely on the their
respective template strands. These 'long products' are
shown immediately above and below the dsDNA.
3' direction. Both produce 'long
products' as they extend indefinitely on the their
respective template strands. These 'long products' are
shown immediately above and below the dsDNA.
The second denaturation again separates the two strands of the target DNA, with the same result as in the first cycle. The 'long products' from the first cycle have incorporated into their 5' ends the sequences of the Forward or Reverse Primers (shown below and above the long products, respectively). The second annealing allows the Forward (5'-F-3') and Reverse (3'-R-5') primers to anneal to their complementary sequences on the long products. These second extensions then proceeds just until they reach the 5' ends of their respective long products. For the first time, the rxn produces ssDNA products exactly as long as the distance between the 5' ends of the two primers. These 'unit length products' are shown immediately above and below the two 'long products'.
The third denaturation separates all previously paired strands. The third annealing allows the Forward and Reverse primers to anneal to the 3' complementary ends of the two ssDNA 'unit length products'. For the first time, extension of these primers produces dsDNA products of unit length, with 3' ends complementary to the Forward primer on one strand, and the Reverse primer on the other. The PCR process has made the regions under the primers at either end of the dsDNA product identical to the primer sequences: naturally occurring SNPs cannot be detected in those regions.
On the fourth and subsequent cycles, the dsDNA PCR products are denatured, Forward and Reverse primers anneal to the 3' ends of either strand, and new strands are synthesized off each template to the limits of the primer. The number of PCR products doubles each cycle: replication of the original template DNA and the 'long products' increases only linearly. The final PCR product is almost entirely the unit length product.
That is, exponential replication of the DNA target begins only of the fourth PCR cycle. Compare this with the simplified diagram, which shows the process as though it works beginning in the first round of amplification.
The first denaturation step separates the two strands. The first Annealing step allows the Forward (5'F3') and and Reverse (3'R5') Primers to anneal to their complementary sequences in the top and bottom strands, respectively. The primers are on opposite strands at either end of the intervening target region. The 5'
The second denaturation again separates the two strands of the target DNA, with the same result as in the first cycle. The 'long products' from the first cycle have incorporated into their 5' ends the sequences of the Forward or Reverse Primers (shown below and above the long products, respectively). The second annealing allows the Forward (5'-F-3') and Reverse (3'-R-5') primers to anneal to their complementary sequences on the long products. These second extensions then proceeds just until they reach the 5' ends of their respective long products. For the first time, the rxn produces ssDNA products exactly as long as the distance between the 5' ends of the two primers. These 'unit length products' are shown immediately above and below the two 'long products'.
The third denaturation separates all previously paired strands. The third annealing allows the Forward and Reverse primers to anneal to the 3' complementary ends of the two ssDNA 'unit length products'. For the first time, extension of these primers produces dsDNA products of unit length, with 3' ends complementary to the Forward primer on one strand, and the Reverse primer on the other. The PCR process has made the regions under the primers at either end of the dsDNA product identical to the primer sequences: naturally occurring SNPs cannot be detected in those regions.
On the fourth and subsequent cycles, the dsDNA PCR products are denatured, Forward and Reverse primers anneal to the 3' ends of either strand, and new strands are synthesized off each template to the limits of the primer. The number of PCR products doubles each cycle: replication of the original template DNA and the 'long products' increases only linearly. The final PCR product is almost entirely the unit length product.
That is, exponential replication of the DNA target begins only of the fourth PCR cycle. Compare this with the simplified diagram, which shows the process as though it works beginning in the first round of amplification.
Figure & text ©2024 by Steven M. Carr