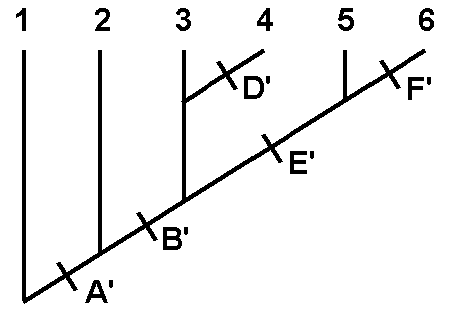
Fig. 1. Cladogram indicating phylogenetic relationships among taxa 1- 6.
Laboratory 4 - CLADISTIC ANALYSIS
In preparation for your lab read, pp. 26 - 30 and 437 - 446 of your text, Evolutionary Analysis Second Edition by Freeman and Herron, paying particular attention to Box 13.1 and Figure 13.9. [see also Bio2900 webpage for Topic 11]
Cladistic analysis consists of three processes: selection of characters, coding of characters and development of a cladogram to explain the evolutionary relationships among the taxa. The techniques identify monophyletic groups on the basis of “shared derived characters” or synapomorphies. The smallest of these groups form the “twigs” of a branching diagram. They are joined to larger branches by identifying “nested” synapomorphies, i.e. each set of like characters which unites closely related taxa, is a subset of those characters which identify a larger monophyletic group. By continuing this process, cladistic methods result in diagrammatic phylogenetic hypotheses (cladograms). Because the number of characters and character changes increases dramatically with the addition of each taxon computer methods are most often used to generate and evaluate the many possible cladograms.
The taxa under consideration may be compared with an outgroup which determines the polarity, or direction of character change, as well as rooting the branching diagram. However the analysis is constrained by these assumptions. Unconstrained, simultaneous analysis will often produce a more parsimonious cladogram, i.e. one with fewer evolutionary changes. In this case an outgroup comparison is not made and polarity is determined from the resulting cladogram. However this is more difficult to achieve without the aid of computer analysis.
In this lab you will use a few taxon only, to become familiar with basic cladistic methods. Specifically, you will use a subset of a group of well known hypothetical organisms. Each pair of students will be assigned “specimens” of plants or animals. The hypothetical class of plants called “Dendrogrammaceae” was created by W. H. Wagner Jr. (1972). The animal-like Caminalcules were created by J. H. Camin (1965). You will use the following cladistic methods to create a phylogenetic tree:
(1) You will be provided with a hypothetical ancestor (outgroup) to your particular group of organisms. You will also be provided with a list of characters OR asked to choose a specified number of appropriate characters which will form the basis of your analysis.
(2) Observe all taxa using the list of characters and character states. Determine the plesiomorphic state and subsequent changes for each character. Code the characters using the Wagner method, which designates the plesiomorphic character state as 0, with subsequent changes coded as 1, 2 etc. Ideally you should use only qualitative characters e.g. petals, no petals. If you choose quantitative characters you divide them into categories so that they can be coded as qualitative characters e.g. Body length < 2 cm = 0, 2 cm - 4 cm = 1, > 4 cm = 2. . Construct Table 1, to summarize the data for the taxa, as in the example on the next page.
(3) Construct Table 2, “A Matrix of Shared Derived Characters”, as in the example on the next page. Note that a taxon with a character state coded “2" is assumed to have accumulated 2 evolutionary changes for this trait, having undergone change “1" previous to change “2".
(4) Using the hypothetical organisms and the information provided in tables 1 and 2, construct the best cladogram to explain the distribution of characters across the taxa. You may need to construct several cladograms suggested by the data. Evaluate these using the principle of parsimony. Present your final cladogram using Figure 13.1 and 13.9 from Freeman and Herron (2001) as a guide. As in Figure 13.9, use numbers or letters to indicate the evolutionary changes. Include a key to identify the changes at the bottom of the page.
Before your lab period, complete the following practice exercise and be prepared to pass in / present your tables and cladograms.
Prelab practice exercise.
Consider an hypothesized data set consisting
of 6 taxa and 6 characters A - F, each with character states
coded 0 and 1.
Taxon # 1 is an outgroup which possesses
the plesiomorphic state, coded “0", for all six characters.
The characters and character states have been identified for each taxon and the results are summarized in Table 1.
Table 1: Data Matrix indicating the character state for selected characters for taxa 1 - 6.
Character Taxa
#1 #2 #3 #4
#5 #6
A
0 1 1
1 1 1
B
0 0 1
1 1 1
C
0 0 0
0 0 0
D
0 0 0
1 0 0
E
0 0 0
0 1 1
F
0 0 0
0 0 1
The data in Table 1 are then compared to determine which taxa share apomorphic (derived) states for the characters under consideration. The results are summarized in Table 2.
Table 2: Matrix of shared derived characters for taxa 1-6.
Taxa 1
2 3 4 5 6
1
- 0 0 0 0 0
2
- 1 1 1 1
3
- 2 2 2
4
- 2 2
5
- 3
6
-
From the data in Table 2, taxa 5 & 6 share the greatest number of derived characters.
Referring to Table 1, they are A' , B' and E'. The taxa differ by F' in taxon 6.
Refer to Tables 2 & 1 again. Both taxa 3 & 4 have two derived traits, A' & B', in common with taxa 5 & 6, and differ by possessing the plesiomprphic state of E. They differ from one another by D' in taxon 4.
Taxon 2 has one derived character,
A',
in common with all previously considered taxa and differs from all by having
the plesiomorphic state for character B.
This analysis is used to construct Figure 1. Begin the branching diagram at the most distant twigs (the taxa with the greatest number of shared derived traits), as in the previous discussion.

Fig. 1. Cladogram indicating phylogenetic
relationships among taxa 1- 6.
Consider the addition of taxon #7, with
characters A', B', C', D', E and F.
[correction: not E' and F'
]
Before coming to lab, reconstruct Tables 1 & 2 and the cladogram to include taxon #7.