Bio4241 -
Comprehensive exam questions, for Tuessday,
07 April 2009, two hours between 10-1P
Instructions: Prepare answers for
each of the following questions: your answer for each should fit on the
front and back of one lined sheet of paper. For the exam, I will select
at random three question for
Part I, and three questions
from Part II. You will answer any two questions
from each part; you may not answer a question pertaining to your own
group's presentation.
You may bring notes or outlines of your prepared answers, on the front
and back of one 8.5 x 11 sheet of paper. [Please don't try to cram complete essays
onto this page!]. You will have two hours to answer the four questions.
I. Questions
based
on research papers
What is a Gene:
How would you define the concept "gene"
in 2009? Compare and contrast your definition with the gene as it was
understood by the time of the “cracking of the genetic code” ca. 1965,
and the classical notion of "gene" as it was before Watson & Crick
(1953).
S.
Suppose that the Luria & Delbruck
(1941)
thought experiment #2 [http://www.mun.ca/biology/scarr/4241smc_Luria-Delbruck_TE2.html]
were continued for two more generations, and that mutation occurs in
one cell of culture #3 in the next-to-last generation. Calculate the
expected mean and variance in the final generation, and the mutation
rate.
1. Consider
Experiments 5
& 6
of Hershey
& Chase (1951). Given what we now know about the functions
of DNA and protein in phage infection, what would be the ideal
numerical results expected if the experiment performed exactly as
expected? Give some reasons why the observed results differs from this
expectation. [http://philos.biol.mun.ca/%7Eb4241/4241_kmf/4241kmfexp3.html].[http://philos.biol.mun.ca/%7Eb4241/4241_kmf/4241_kmf_point6.html]
2. Sketch
the expected results from the Meselson-Stahl
(1958) experiment, [http://philos.biol.mun.ca/~b4241/b4241_bank/bandresults_4241.html],
if DNA replication were (1) conservative or (2) dispersive. Provide two
alternative modes for dispersive replication, and show the different
results expected for each.
3.
After Nirenberg & Matthaei (1961)
began to “crack the genetic code,” Khorana polymerized UG, UGG,
or UGGG di, tri, and
tetra-nucleotides into longer polynucleotides. The various
polynucleotides produced different mixture of polypeptides. Explain the
logic by which he was able to infer the genetic code for the
triplets
produced by these experiments. [http://www.mun.ca/biology/scarr/di-tri-tetraribonucleotides.html]
5.
King & Wilson (1975) used
a series of calculations to whos that “Chimps
and humans are 99% similar.” Summarize the calculations used by to measure
the genetic similarity of these two species.
6.
With respect to Sanger sequencing,
compare and contrast autoradiographic
and laser fluorometric
methods of detecting the results of dideoxy-sequencing reactions.
8.
With respect to Saiki et al.’s (1988)
PCR method, sketch and explain the nature of
amplification products obtained in the first three cycles of a PCR
experiment. How does this differ from simple explanations given in
Bio2250?
II. Questions
based on book chapters
Chap. 12: Distinguish between "Forward"
and "Reverse" genetics.
Describe how each approach would be applied to understanding the nature
of wrinkled vs. round peas.
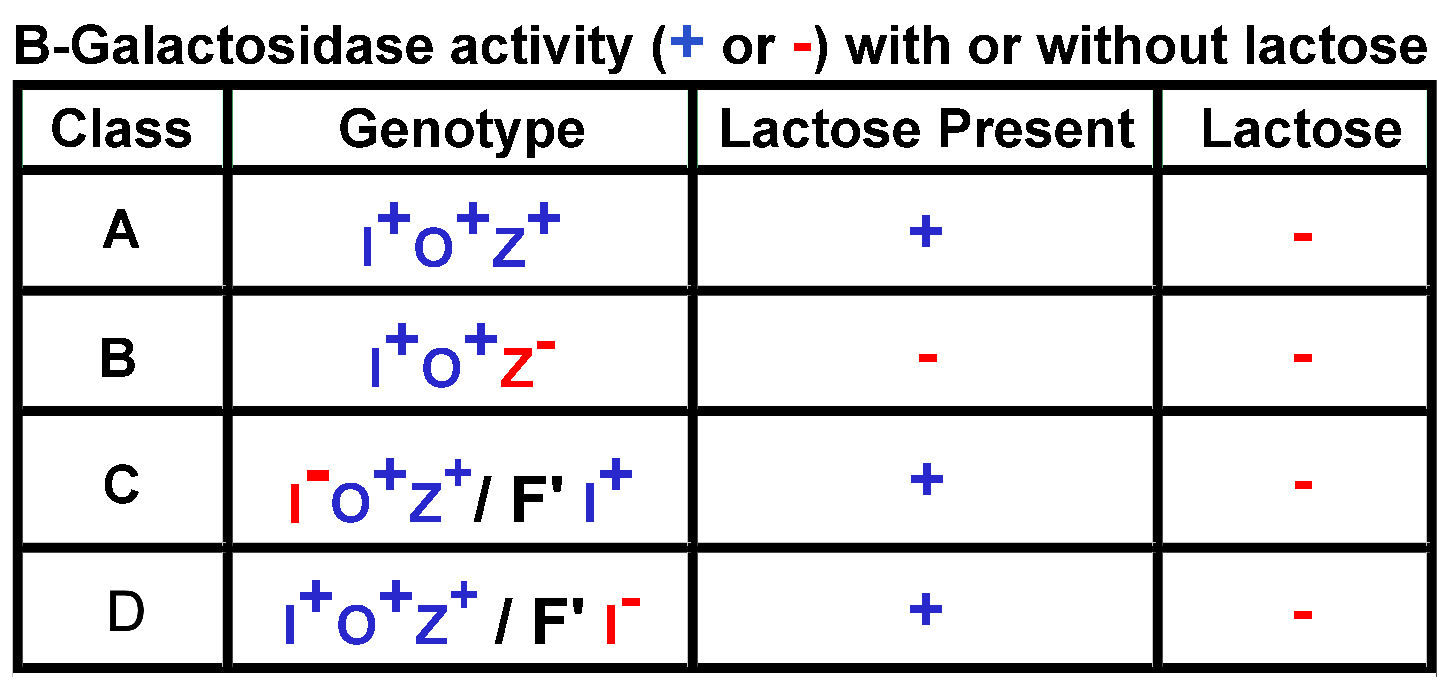
Chap.
13: For each of the four lac-operon genotype classes in the
table below,
explain the associated phenotypic pattern of B-Galactosidase activity,
in the presence of absence of lactose.
Chap. 14: Answer question
15.21 (p. 508), concerning the genetics of Retinoblastoma.
Chap. 16: Compare and contrast
chromosomal mechanisms of sex determination in Drosophila and humans.
Chap. 16: Explain in detail the genetic and
technical basis of Figure b at
[http://philos.biol.mun.ca/~b4241/4241_aapr16b/gap_pairrule.html].
Why are there stripes? How were the stripes visualized?

