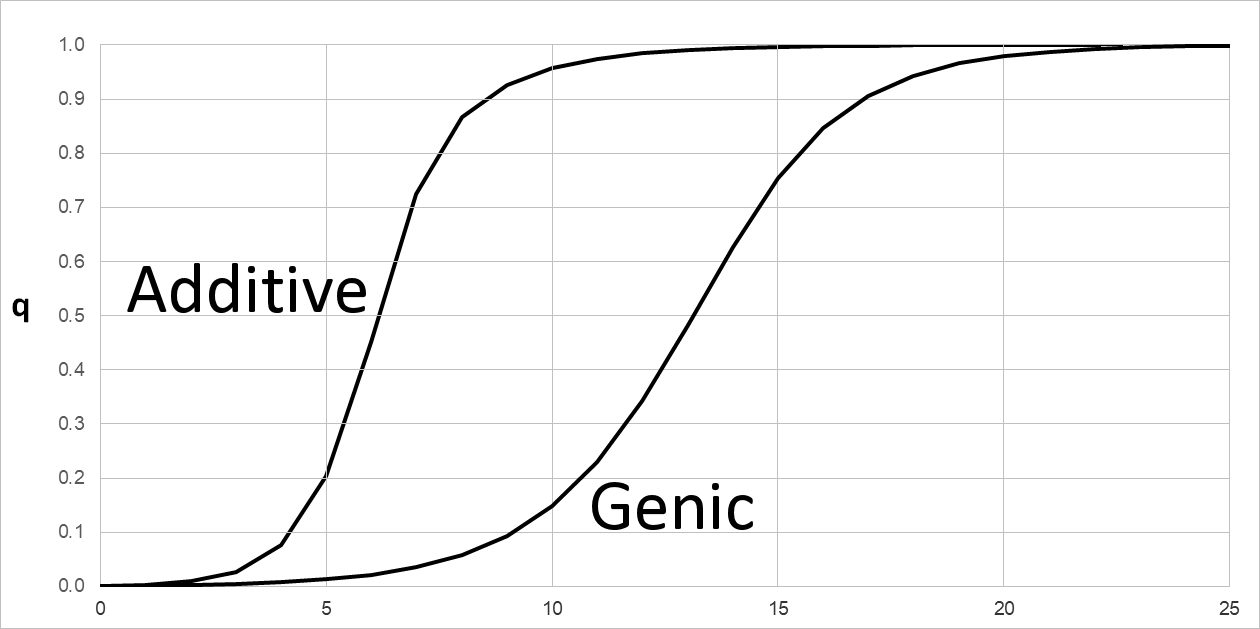
Natural Selection on phenotypes with Additive & Genic fitness
In classical genetics, if the phenotype
of the AB genotype is precisely intermediate
between those of the two homozygous genotypes AA and
BB, the A and B alleles are
described as semi-dominant.
If the phenotype of the AB genotype is
intermediate between AA & BB, but
closer to that of AA than the BB, A is
described as incompletely dominant
to B. If AB is closer to BB, then B
is the incompletely dominant allele. Note once again that, if B is
incompletely dominant to A, it is not because
B has superior phenotype (and might be said to "dominate"
the other allele in its effect), nor is it because f(B)
> f(A) (and might be said to "predominate"
the other), but because the phenotype of the AB
genotype is intermediate between that
of the AA and BB. Genetic dominance is a
genotypic, not a phenotypic, relationship.
The graph shows semi-dominance, as it occurs in the Additive fitness model. Let q0 = 0.001. If the selection coefficient s = 0.4, then WBB = (1), WAB = (1 - s), and WAA = (1 - 2s), and fitness values are so WBB = 1.0, WAB = 0.6, and WAA = 0.2. Each A allele contributes an additive selective disadvantage of s = 0.4, so that an AA homozygote is at twice the disadvantage of the AB heterozygote. Note that for s < 0.5, WAA < 0, which is undefined. One convention is to round these fitness to 0.0.
Compare this with the model for Genic fitness, also called multiplicative fitness. Let q0 = 0.001. With s = 0.4 as above, WBB = (1), WAB = (1 - s), and WAA = (1 - s)(1 - s) = (1 - s)2 , so WBB = 1.0, WAB = 0.6, and WAA = 0.36. That is, each A allele reduces fitness by a factor of (1 - s). In either model, the fitness effect of a single allele in the heterozygote is (1 - s). However, the two models make very different predictions about fitness in the range 0.1 ~ s ~ 0.5: in the example, WAA = 0.20 & 0.36 in the Additive and Genic models, respectively. At smaller values of s, the difference between models becomes negligible. When s << 0.1 such that s2 << 2s, genic fitness (1 - s)2 = 1 - 2s + s2 ~ (1 - 2s) as in additive fitness.
Simple additive dominance may be typical at many gene loci, where the phenotype is a consequence of equal expression by both alleles. Extending the classical genetic model to protein-coding loci, each allele would contribute half the total amount of gene product. For example, so-called "null alleles" protein electrophoresis occur when one non-functional allele produces no product. For a monomeric protein, only one band is seen. The other, functional allele produces 50% of the expected gene product, which may (or may not) provide sufficient enzyme product for standard phenotypic expression. Alternatively, the standard allele may be "up-regulated" so that the amount of gene product expressed in the AB heterozygote is (much) closer to that of the AA homozygote.
Incomplete genic dominance may be typical at other gene loci, where the effect of alleles on the phenotype is multiplicative. For example, if the products of a single locus compete for binding to the two sites of an enzyme, the chance that Site 1 AND Site 2 will both bind the less functional product is a product, not a sum.
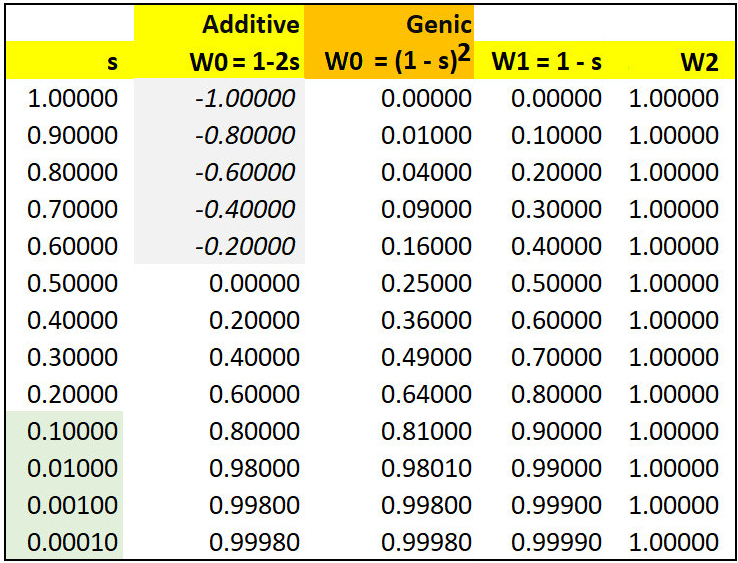
It remains a major point of contention what fraction of heterozygous allelic variation detected originally by protein electrophoresis and (or) nowadays by DNA sequencing has any measurable effect on the observed phenotype relative to that of the homozygotes, as is clear from the table below. The so-called "Neutralist - Selectionist" controversy will be discussed elsewhere in the course.
The graph shows semi-dominance, as it occurs in the Additive fitness model. Let q0 = 0.001. If the selection coefficient s = 0.4, then WBB = (1), WAB = (1 - s), and WAA = (1 - 2s), and fitness values are so WBB = 1.0, WAB = 0.6, and WAA = 0.2. Each A allele contributes an additive selective disadvantage of s = 0.4, so that an AA homozygote is at twice the disadvantage of the AB heterozygote. Note that for s < 0.5, WAA < 0, which is undefined. One convention is to round these fitness to 0.0.
Compare this with the model for Genic fitness, also called multiplicative fitness. Let q0 = 0.001. With s = 0.4 as above, WBB = (1), WAB = (1 - s), and WAA = (1 - s)(1 - s) = (1 - s)2 , so WBB = 1.0, WAB = 0.6, and WAA = 0.36. That is, each A allele reduces fitness by a factor of (1 - s). In either model, the fitness effect of a single allele in the heterozygote is (1 - s). However, the two models make very different predictions about fitness in the range 0.1 ~ s ~ 0.5: in the example, WAA = 0.20 & 0.36 in the Additive and Genic models, respectively. At smaller values of s, the difference between models becomes negligible. When s << 0.1 such that s2 << 2s, genic fitness (1 - s)2 = 1 - 2s + s2 ~ (1 - 2s) as in additive fitness.
Simple additive dominance may be typical at many gene loci, where the phenotype is a consequence of equal expression by both alleles. Extending the classical genetic model to protein-coding loci, each allele would contribute half the total amount of gene product. For example, so-called "null alleles" protein electrophoresis occur when one non-functional allele produces no product. For a monomeric protein, only one band is seen. The other, functional allele produces 50% of the expected gene product, which may (or may not) provide sufficient enzyme product for standard phenotypic expression. Alternatively, the standard allele may be "up-regulated" so that the amount of gene product expressed in the AB heterozygote is (much) closer to that of the AA homozygote.
Incomplete genic dominance may be typical at other gene loci, where the effect of alleles on the phenotype is multiplicative. For example, if the products of a single locus compete for binding to the two sites of an enzyme, the chance that Site 1 AND Site 2 will both bind the less functional product is a product, not a sum.
It remains a major point of contention what fraction of heterozygous allelic variation detected originally by protein electrophoresis and (or) nowadays by DNA sequencing has any measurable effect on the observed phenotype relative to that of the homozygotes, as is clear from the table below. The so-called "Neutralist - Selectionist" controversy will be discussed elsewhere in the course.

Table & text material © 2025 by Steven M. Carr